For the first time, a yeast strain with a genome made up of more than 50% synthetic DNA has been created in the lab. This is the next step in a 15-year project to design and construct a totally synthetic eukaryotic genome from scratch, which the team aims to complete this year.
The synthetic yeast genome project, also known as Saccharomyces cerevisiae (Sc) 2.0, is made up of researchers from all over the world who have individually assumed responsibility for creating a synthetic version of one of the yeast genome’s 16 chromosomes.
‘We have roughly ten teams from prominent universities on four continents; it’s a truly worldwide effort,’ says Patrick Cai, chair in synthetic genomics at the University of Manchester and the Sc2.0 project’s international coordinator.
According to Ben Blount, the leader of one of the UK-based teams, yeast has been used as a model organism for eukaryotes for many years, so scientists likely know more about yeast than any other creature. He describes the yeast genome as “really nice and compact.” ‘It also has this amazing intrinsic capacity to simply glue DNA together. So, if you have two DNA sequences that are identical, the yeast will just sew that DNA together.’
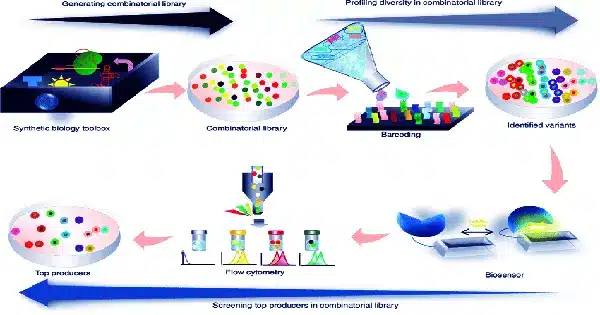
A collection of eight studies published lately in Cell, Molecular Cell, and Cell Genomics presented the Sc2.0 consortium’s synthetic yeast chromosomes and emphasized crucial traits they discovered. Furthermore, the consortium released a proof-of-concept study in which 6.5 of the synthetic chromosomes, plus an extra ‘neochromosome’ made up entirely of transfer RNA (tRNA) genes, were combined into a single cell capable of surviving and replicating in a manner comparable to wild yeast.
‘We demonstrate that we have the technology to integrate over 50% of the synthetic DNA in a single cell,’ Cai says. ‘This is a watershed moment in biology and biotechnology; this is the most synthetic organism we’ve ever created.’
Blount, whose team was in charge of chromosome XI, adds that the design of each chromosome was meticulously controlled by a set of rigid design rules before an expensive and lengthy ‘debugging’ procedure. Instead of being a straight copy of the natural genome, the Sc2.0 synthetic genome contains thousands of designer edits, such as deletion of mobile elements and introns, relocation of transfer RNAs (tRNAs), and stop codon swapping, to increase genome stability and generate a variety of phenotypes with desirable properties, such as virus immunity.
Blount’s team also demonstrated that its chromosome may be used as a new approach for investigating extrachromosomal circular DNAs. These free-floating DNA loops that loop out of the genome are becoming more well-known for their significance in aging and cancer.2 ‘These rings are methods for tumors to become resistant to many of the medicines we use,’ explains Blount.
Cai’s lab also constructed a fully synthetic ‘neochromosome’ composed entirely of 275 tRNA genes transplanted from the original chromosomes.3 ‘There is nothing like this in nature,’ adds Cai. ‘However, we implanted it in the yeast [cell], and it appears to be happy with it, indicating that it is working.’
The process of bringing all of the chromosomes together within a single yeast cell is now beginning, and the whole synthetic genome is expected to be completed within a year.
The Sc2.0 work, according to John Glass, leader of the synthetic biology group at the J. Craig Venter Institute, where a team generated a bacterial cell with a synthetic genome in 2010. ‘With each new step closer to their goal of creating a yeast cell with a completely synthetic genome, they discover extraordinary and unexpected things about eukaryotic biology that are fascinating and, over time, will have extraordinary consequences for medicine, industry, and our understanding of ourselves,’ he explains.
‘They have kept this group of at least 16 different academic, industrial, and government organizations together for more than a decade, and they have kept it funded from a wide range of sources. And this is not a little task,’ he continues.