According to researchers at Tokyo Tech, in-cell engineering can be a powerful tool for synthesizing functional protein crystals with promising catalytic properties. The researchers created hybrid solid catalysts for artificial photosynthesis using genetically modified bacteria as an environmentally friendly synthesis platform. These catalysts have high activity, stability, and durability, demonstrating the potential of the proposed novel approach.
Protein crystals, like regular crystals, are well-ordered molecular structures with a wide range of properties and a great deal of flexibility. They can naturally assemble from materials found within cells, which not only reduces synthesis costs but also has a lower environmental impact.
Although protein crystals are promising as catalysts due to their ability to host a wide range of functional molecules, current techniques only allow for the attachment of small molecules and simple proteins. Thus, it is critical to develop methods for producing protein crystals containing both natural enzymes and synthetic functional molecules in order to fully exploit their potential for enzyme immobilization.
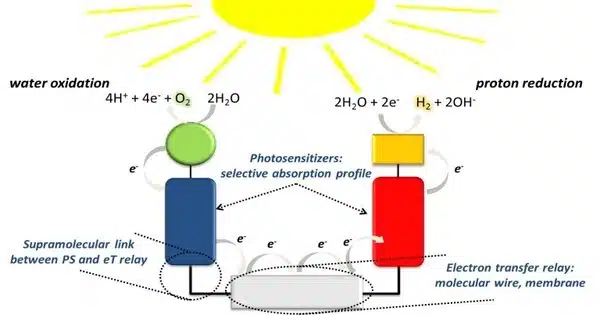
Researchers developed an innovative strategy for producing hybrid solid catalysts based on protein crystals. Their approach combines in-cell engineering and a simple in vitro process to produce catalysts for artificial photosynthesis.
In this context, a team of Tokyo Institute of Technology (Tokyo Tech) researchers led by Professor Takafumi Ueno has developed an innovative strategy for producing hybrid solid catalysts based on protein crystals. Their approach combines in-cell engineering and a simple in vitro process to produce catalysts for artificial photosynthesis, as explained in their paper published in Nano Letters.
The building block of the hybrid catalyst is a protein monomer derived from a virus that infects the Bombyx mori silkworm. The researchers introduced the gene that codes for this protein into Escherichia coli bacteria, where the produced monomers formed trimers that, in turn, spontaneously assembled into stable polyhedra crystals (PhCs) by binding to each other through their N-terminal α-helix (H1). Additionally, the researchers introduced a modified version of the formate dehydrogenase (FDH) gene from a species of yeast into the E. coli genome. This gene caused the bacteria to produce FDH enzymes with H1 terminals, leading to the formation of hybrid H1-FDH@PhC crystals within the cells.
The team extracted the hybrid crystals out of the E. coli bacteria through sonication and gradient centrifugation, and soaked them in a solution containing an artificial photosensitizer called eosin Y (EY). As a result, the protein monomers, which had been genetically modified such that their central channel could host an eosin Y molecule, facilitated the stable binding of EY to the hybrid crystal in large quantities.
The team was able to create highly active, recyclable, and thermally stable EY•H1-FDH@PhC catalysts that, when exposed to light, can convert carbon dioxide (CO2) into formate (HCOO), mimicking photosynthesis. Furthermore, when compared to the free enzyme, they maintained 94.4% of their catalytic activity after immobilization.
“The proposed hybrid crystal’s conversion efficiency was an order of magnitude higher than that of previously reported compounds for enzymatic artificial photosynthesis based on FDH,” Prof. Ueno emphasizes. “Moreover, after undergoing both in vivo and in vitro engineering processes, the hybrid PhC remained in the solid protein assembly state, demonstrating the remarkable crystallizing capacity and strong plasticity of PhCs as encapsulating scaffolds.”