The researchers created and tested a new method for high-throughput single-cell sorting that use stimulated Raman spectroscopy rather than the usual approach of fluorescence-activated cell sorting. The novel method could provide a label-free, nondestructive method of sorting cells for a range of applications such as microbiology, cancer detection, and cell therapy.
The researchers created and tested a new method for high-throughput single-cell sorting that use stimulated Raman spectroscopy rather than the usual approach of fluorescence-activated cell sorting. The novel method could provide a label-free, nondestructive method of sorting cells for a range of applications such as microbiology, cancer detection, and cell therapy.
Jing Zhang from Boston University will present this research at Frontiers in Optics + Laser Science (FiO LS), which will be held 9 – 12 October 2023 at the Greater Tacoma Convention Center in Tacoma (Greater Seattle Area), Washington.
Our approach (stimulated Raman-activated cell ejection, S-RACE) offers an innovative way to sort cells based on their intracellular chemical composition in a high-throughput manner.
Jing Zhang
“Our approach (stimulated Raman-activated cell ejection, S-RACE) offers an innovative way to sort cells based on their intracellular chemical composition in a high-throughput manner,” Zhang explains.
“The separated cell populations could be subjected to various downstream phenotypic and/or genomic analyses.” Additionally, its affinity with tiny cells makes it useful for separating bacteria and other microbes. Pathogens or cells with certain metabolic profiles, for example, might be immediately captured from their natural habitat, such as water bodies, soil, or the gastrointestinal system, using S-RACE. Following sequencing allows for activities like cell taxonomic identification and ecological function assessment.”
Flow cytometry is used in many biomedical fields to rapidly count and characterize various types of cells, including blood cells, stem cells, cancer cells and microorganisms. Sorting cells based on their size, granularity or expression of cell surface and intracellular molecules can be used to gain insights into biological processes or to separate out cells with certain characteristics for additional analysis.
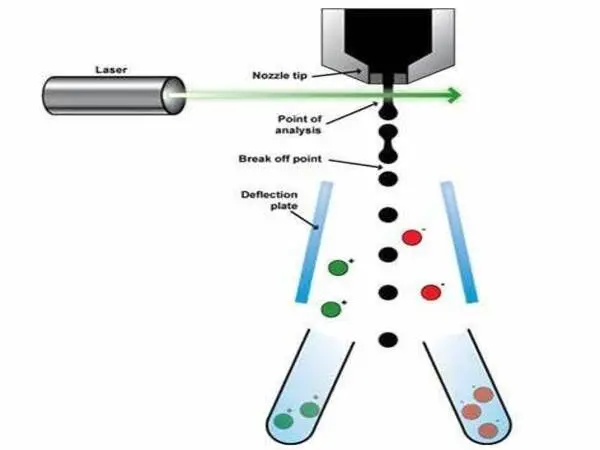
Although most current high-throughput cell sorting methods rely on fluorescence signals for sorting, fluorescence labels can disturb cell function and can’t be used with small molecules. Raman spectroscopy is a promising alternative because it offers label-free and non-destructive single-cell measurement by obtaining a chemical fingerprint of the cell. However, it has been difficult to achieve both a strong Raman signal and a practical microfluidic setup for imaging cells.
The researchers describe how they surmounted this obstacle by employing stimulated Raman spectroscopy, which gives a signal that is many orders of magnitude stronger than the more often employed spontaneous Raman scattering. To sort, stimulated Raman images are obtained to identify objects or cells of interest, and then a 532-nm pulsed laser is directed at the cell using 2D galvo mirrors. Finally, an acousto-optic modulator serves as a rapid pulse picker, allowing single laser pulses to drive the chosen cell into the collector. Each ejection only takes around 8 milliseconds.
The researchers first showed their stimulated Raman-activated cell ejection method with a combination of 1-micron polymer beads, reaching approximately 95% purity and 98% throughput with approximately 14 ejections conducted per second. They also demonstrated that the procedure could be applied to fixed bacteria.
To apply the sorting approach to live yeast cells, the researchers utilized an agar plate as a collector and added a thin layer of agar to the ejection module to shield the cells from heat and drying. The researchers utilized the apparatus to eject roughly 340 yeast cells, and after about 40 hours, they saw good cell growth in the receiving plate. They also showed that other genomic analysis approaches such as quantitative polymerase chain reaction could be integrated with the sorting approach.













