Cells must be able to precisely control which of their genetic material’s many genes they use. This is done in transcription factories, which are molecular clusters in the nucleus. The formation of transcription factories now resembles the condensation of liquids, according to researchers. Their findings will contribute to a better understanding of disease causes and the advancement of DNA-based data storage systems.
Cells must be able to precisely control which of their genetic material’s many genes they use. This is done in transcription factories, which are molecular clusters in the nucleus. Researchers from the Karlsruhe Institute of Technology (KIT), Friedrich-Alexander-Universität Erlangen-Nuremberg (FAU), and the Max Planck Institute for Physics and Medicine (MPZPM) discovered that the formation of transcription factories is similar to liquid condensation. Their findings will contribute to a better understanding of disease causes and the advancement of DNA-based data storage systems. The findings were published in the journal Molecular Systems Biology.
Our findings demonstrate how the biological cell organizes such processes in a timely and consistent manner. Our computer simulations and functional concepts can be directly transferred to artificial DNA systems and used to support their design.
Lennart Hilbert
There are over 20,000 different genes in human genetic material. However, each cell only uses a small portion of the information stored in the genome. As a result, cells must precisely control which genes they use. Cancer or an embryonic growth disorder may develop if this is not done. Transcription factories are essential in the selection of active genes. “These factories are nucleoplasmic molecular clusters that combine the correct selection of active genes and the read-out of their sequence at a central location,” Lennart Hilbert explains.
The Junior Professor for Systems Biology/Bioinformatics at the Zoological Institute (ZOO) of KIT also heads a working group at KIT’s Institute of Biological and Chemical Systems, Biological Information Processing (IBCS-BIP).
Setup and Start within a Few Seconds
Cellular and molecular biologists have been studying how transcription factories are set up and activated in a matter of seconds for decades. So far, the results indicate that only processes known from industrial and technical polymer and liquid materials are relevant. The current focus of research is on phase separation as a central mechanism. When separating oil from water in everyday life, phase separation can be observed. However, it is unclear how phase separation contributes to the establishment of transcription factories in living cells.
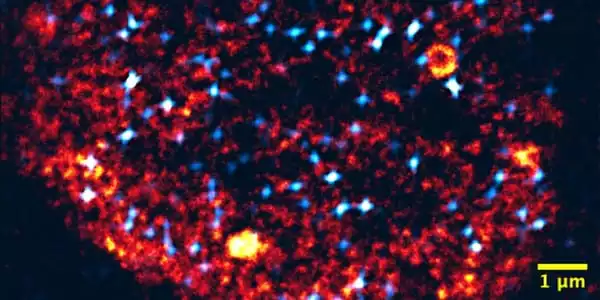
Researchers from KIT’s Institute of Biological and Chemical Systems (IBCS), Zoological Institute (ZOO), Institute of Applied Physics (APH), and Institute of Nanotechnology (INT), in collaboration with scientists from Erlangen’s FAU and MPZPM, as well as the University of Illinois at Urbana-Champaign/USA, have discovered new information about the formation of transcription factories: It is analogous to liquid condensation. This was published in the journal Molecular Systems Biology. Agnieszka Pancholi of IBCS-BIP and ZOO, and Tim Klingberg of FAU and MPZPM, are the first co-authors.
Latest Light Microscopy Combined with Computer Simulations
The researchers note in their paper that condensation to form transcription factories resembles steamy glasses or windows. Only in the presence of a receptive surface does liquid condensate, and then very quickly. As condensation surfaces in living cells, specially marked areas of the genome are used. The liquid-coated areas allow relevant gene sequences and additional molecules to adhere, eventually activating the adhering genes.
These results were obtained through interdisciplinary collaboration. The latest light microscopes developed by Professor Gerd Ulrich Nienhaus’ Chair at APH were used to study zebrafish embryos. These findings were then linked to computer simulations at the FAU Chair for Mathematics, which is led by Professor Vasily Zaburdaev. The combination of observations and simulations makes the condensation process reproducible and explains how living cells can quickly and reliably set up transcription factories.
A new understanding of condensed liquids in living cells has recently resulted in completely new approaches to treating cancer and nervous system diseases. Startups developing new drugs are now pursuing these approaches. Other research efforts are centered on the use of DNA sequences as digital data storage systems.
Meanwhile, several working groups have demonstrated the basic feasibility of DNA-based data storage systems. Reliable data storage and read-out in such DNA storage media remain significant challenges. “Our findings demonstrate how the biological cell organizes such processes in a timely and consistent manner. Our computer simulations and functional concepts can be directly transferred to artificial DNA systems and used to support their design”According to Lennart Hilbert.