Evolution is the process by which populations of organisms change over generations through mechanisms such as natural selection and genetic drift. In natural selection, organisms with traits better suited to their environment are more likely to survive and reproduce, passing those advantageous traits to their offspring. Over time, this can result in the development of new species. Genetic drift, on the other hand, is a random process that can lead to changes in the frequency of alleles (versions of a gene) in a population. These mechanisms, along with mutation (random changes to an organism’s DNA), can cause populations to diverge and evolve into new forms over time.
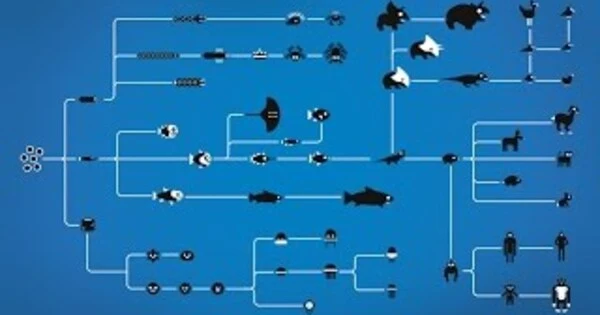
The European mole can easily burrow through the soil thanks to its powerful digging shovels. The same holds true for the Australian marsupial mole. Despite their geographical separation, the two animal species have evolved similar organs – in their case, extremities ideally adapted for digging in the soil.
In such cases, science refers to “convergent evolution,” which occurs when animal and plant species independently develop features with the same shape and function. There are numerous examples: Fish, for example, have fins, as do whales, despite the fact that they are mammals. Birds and bats have wings, and many creatures, from jellyfish to scorpions to insects, have evolved the same weapon when it comes to using poisonous substances to defend themselves against attackers: the venomous sting.
We have developed a novel metric of molecular evolution that can accurately represent the rate of convergent evolution in protein-coding DNA sequences.
Dr. Kenji Fukushima
Identical characteristics despite lack of relationship
Scientists around the world are clearly interested in determining which changes in the genetic material of the respective species are responsible for the fact that identical characteristics have evolved in them despite the fact that they have no relationship.
This is proving difficult to find: “Such traits — we call them phenotypes — are, of course, always encoded in genome sequences,” says Dr. Kenji Fukushima of the Julius-Maximilians-Universität (JMU) Würzburg. Mutations, or changes in genetic material, can be the catalyst for the emergence of new traits.
However, because the underlying mutations are largely random and neutral, genetic changes rarely result in phenotypic evolution. As a result, a massive number of mutations accumulate over the extreme time scales at which evolutionary processes occur, making detecting phenotypically significant changes extremely difficult.
Novel metric of molecular evolution.
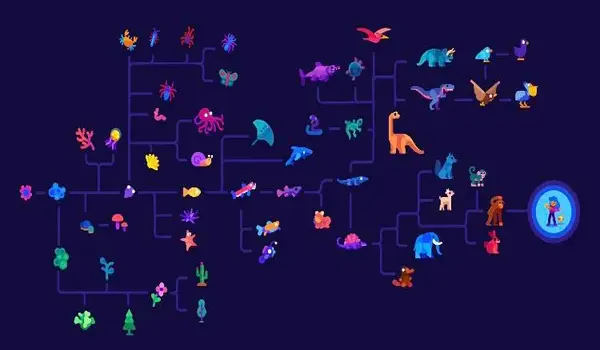
Fukushima and his colleague David D. Pollock of the University of Colorado (USA) have now succeeded in developing a method that produces significantly better results than previous methods in the search for the genetic basis of phenotypic traits. They describe their approach in the current issue of the journal Nature Ecology & Evolution.
“We have developed a novel metric of molecular evolution that can accurately represent the rate of convergent evolution in protein-coding DNA sequences,” says Fukushima, describing the main result of the now-published work. This new method, he claims, can reveal which genetic changes are linked to organism phenotypes over hundreds of millions of years of evolution. It thus offers the possibility of expanding our understanding of how changes in DNA lead to phenotypic innovations that give rise to a great diversity of species.
Tremendous treasure trove of data as a basis
The fact that in recent years more and more genome sequences of many living organisms across a wide range of species have been decoded and thus made available for analysis forms the foundation of Fukushima and Pollock’s work. “This has enabled us to study the interactions of genotypes and phenotypes on a large scale at a macroevolutionary level,” Fukushima says.
However, because many molecular changes are nearly neutral and have no effect on any traits, there is often a risk of “false-positive convergence” when interpreting the data, which means that the result predicts a correlation between a mutation and a specific trait that does not exist. Furthermore, such false-positive convergences could be caused by methodological biases.
Correlations over millions of years
“To address this issue, we broadened the framework and created a new metric that measures the error-adjusted convergence rate of protein evolution,” Fukushima says. This allows him to distinguish natural selection from genetic noise and phylogenetic errors in simulations and real-world examples, he claims. The approach, which has been enhanced with a heuristic algorithm, enables bidirectional searches for genotype-phenotype associations, even in lineages that have diverged over hundreds of millions of years, he claims.
The two researchers examined over 20 million branch combinations in vertebrate genes to see how well their metric works. They intend to apply this method to carnivorous plants as a next step. The goal is to discover the genetic basis for these plants’ ability to attract, capture, and digest prey.