Antimicrobial resistance (AMR) poses a threat to the effective prevention and treatment of a growing number of infections caused by bacteria, parasites, viruses, and fungi. AMR occurs when bacteria, viruses, fungi, and parasites evolve and no longer respond to medications, making infections more difficult to treat and increasing the risk of disease spread, severe illness, and death. As a result, the medications become ineffective, and infections remain in the body, increasing the risk of transmission to others.
An international research team has provided important new information about what drives the global spread of genes responsible for antimicrobial resistance (AMR) in bacteria. The collaborative study, led by researchers at the Quadram Institute and the University of East Anglia, brought together experts from France, Canada, Germany, and the United Kingdom and will provide new information to combat the global threat of AMR.
The team discovered that different types of AMR genes varied in their temporal dynamics by examining the whole genome sequences of around two thousand resistant bacteria, mostly Escherichia coli, collected between 2008 and 2016. Some, for example, were discovered in North America and spread to Europe, while others spread from Europe to North America.
Not only did the study look at bacteria from different geographic regions but also from diverse hosts including humans, animals, food (meat), and the environment (wastewater), to define how these separate but interconnected factors influenced the development and spread of AMR. Understanding this interconnectivity embodies the One Health approach and is vital for understanding transmission dynamics and the mechanisms by which resistance genes are transmitted.
We were able to identify the key genes conferring resistance to these critical drugs by assembling such a large and diverse collection of genomes.
Professor Alison Mather
The Joint Programming Initiative on Antimicrobial Resistance (JPIAMR), a global collaboration spanning 29 countries and the European Commission tasked with turning the tide on AMR, funded the study, which was published in the journal Nature Communications. Without global collaboration, AMR will undoubtedly expose millions more people to infections caused by bacteria and other microorganisms that can currently be treated with antimicrobials.
The researchers concentrated on resistance to a specific class of antimicrobials known as extended-spectrum cephalosporins (ESCs). The World Health Organization has classified these antimicrobials as critical because they are a “last resort” treatment for multidrug-resistant bacteria; however, efficacy has declined since their introduction as bacteria have developed resistance.
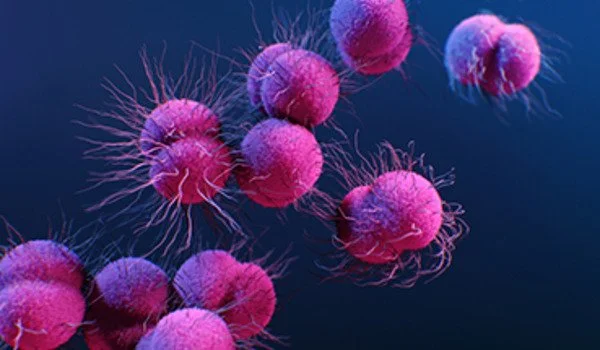
Bacteria that are resistant to ESCs accomplish this by producing specific enzymes known as beta-lactamases, which are capable of inactivating ESCs. These enzymes’ instructions are encoded in genes, specifically two types of genes: extended-spectrum beta-lactamases (ESBLs) and AmpC beta-lactamases (AmpCs).
These genes can be found on bacteria’s chromosomes, where they are passed down to progeny during clonal multiplication, or in plasmids, which are small DNA molecules that exist independently of the bacterium’s main chromosome. Plasmids are mobile and can move between bacteria, providing an alternative method of exchanging genetic material.
This study discovered how some resistance genes spread via clonal expansion of particularly successful bacterial subtypes, while others spread directly on epidemic plasmids across different hosts and countries.
Understanding AMR transmission and the global spread of resistance requires an understanding of the flow of genetic information within and between bacterial populations. This knowledge will help to design critical interventions that can halt AMR in the real world, where bacteria from various hosts and environmental niches interact, and where international travel and trade mean that these interactions are not geographically limited.
Professor Alison Mather of the Quadram Institute and the University of East Anglia stated: “We were able to identify the key genes conferring resistance to these critical drugs by assembling such a large and diverse collection of genomes. We were also able to demonstrate that the majority of resistance to extended-spectrum cephalosporins is spread by a small number of dominant plasmids and bacterial lineages; understanding the mechanisms of transmission is critical for designing interventions to reduce AMR spread.”