Synthetic biology allows cells to be programmed to perform novel functions, such as emitting fluorescent light when they detect a specific chemical. Typically, this is accomplished by modifying cells so that they express genes that can be activated by a specific input.
However, due to the time required for cells to transcribe and translate the necessary genes, there is frequently a long lag time between an event such as detecting a molecule and the resulting output. MIT synthetic biologists have now developed a new method for designing such circuits that is entirely based on fast, reversible protein-protein interactions. This eliminates the need to wait for genes to be transcribed or translated into proteins, allowing circuits to be activated much faster – within seconds.
“We now have a methodology for designing protein interactions that occur on a very short timescale that no one has been able to develop methodically. We’re getting close to being able to engineer any function in a matter of seconds or less “Deepak Mishra, a research associate in MIT’s Department of Biological Engineering and the study’s lead author, says
According to the researchers, this type of circuit could be useful for developing environmental sensors or diagnostics that can detect disease states or impending events such as a heart attack.
Researchers have designed the first synthetic biology circuit that relies exclusively on protein-protein interactions. These circuits can be turned on within seconds, much faster than other synthetic biology circuits.
The study’s senior author, Ron Weiss, is a professor of biological engineering as well as electrical engineering and computer science. It was published today in Science. Tristan Bepler, a former MIT postdoctoral fellow; Bonnie Berger, the Simons Professor of Mathematics and head of the Computation and Biology group in MIT’s Computer Science and Artificial Intelligence Laboratory; Brian Teague, an assistant professor at the University of Wisconsin; and Jim Broach, chair of the Department of Biochemistry and Molecular Biology at Penn State Hershey Medical Center, are also authors.
Protein interactions
Protein-protein interactions are critical steps in many signaling pathways within living cells, including those involved in immune cell activation and responses to hormones or other signals. Many of these interactions involve one protein activating or deactivating another by adding or removing phosphate groups.
The researchers used yeast cells to host their circuit in this study, and they built a network of 14 proteins from yeast, bacteria, plants, and humans. The proteins were modified so that they could regulate each other in the network to produce a signal in response to a specific event.
Their network, the first synthetic circuit made entirely of phosphorylation/dephosphorylation protein-protein interactions, is designed as a toggle switch – a circuit that can quickly and reversibly switch between two stable states, allowing it to “remember” a specific event such as chemical exposure. Sorbitol, a sugar alcohol found in many fruits, is the target in this case.
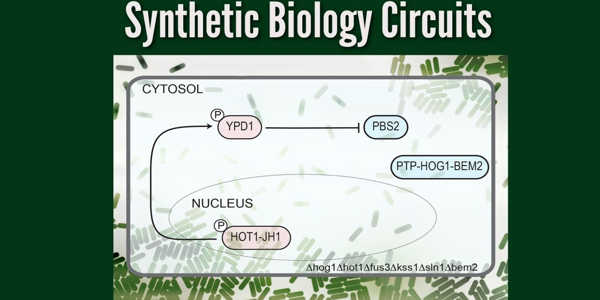
When sorbitol is detected, the cell stores a memory of the exposure in the form of a fluorescent protein that is found in the nucleus. This memory is passed down to future cell generations as well. The circuit can also be reset by exposing it to another molecule, in this case, isopentenyl adenine.
In addition, these networks can be programmed to perform additional functions in response to an input. To demonstrate this, the researchers created a circuit that inhibits cell division when sorbitol is detected.
The researchers can create ultrasensitive sensors that respond to concentrations of the target molecule as low as parts per billion by using large arrays of these cells. The signal can be triggered in as little as one second due to the fast protein-protein interactions. With traditional synthetic circuits, the output could take hours or even days to appear.
“That transition to extremely fast speeds will be really important moving forward in synthetic biology and expanding the types of applications that are possible,” Weiss says.
Complicated networks
The toggle network designed by the researchers in this study is larger and more complex than most synthetic circuits previously designed. After constructing it, the researchers wondered if similar networks existed in living cells. They discovered six previously unknown naturally occurring, complex toggle networks in yeast using a computational model they created.
“We wouldn’t think to look for them because they aren’t obvious. They aren’t always optimal or elegant, but we did come across several examples of such toggle switch behaviors “Weiss explains. “This is a new, engineered-inspired approach to discovering regulatory networks in biological systems.”
The researchers are now hoping to use their protein-based circuits to create sensors that can detect environmental pollutants. Another potential application is the deployment of custom protein networks within mammalian cells to serve as diagnostic sensors within the human body, detecting abnormal hormone or blood sugar levels. In the long run, Weiss hopes to create circuits that can be programmed into human cells to detect drug overdoses or an impending heart attack.
“You could have a situation where the cell reports that information to an electronic device that alerts the patient or the doctor, and the electronic device could also have reservoirs of chemicals that could counteract a system shock,” he says.
The research was supported by the Siebel Scholars Award, an Eni-MIT Energy Research Fellowship, the National Science Foundation Graduate Research Fellowship Program, the Institute for Collaborative Biotechnologies through the U.S. Army Research Office, a National Science Foundation SynBERC grant, and the National Institutes of Health’s Center for Integrated Synthetic Biology.